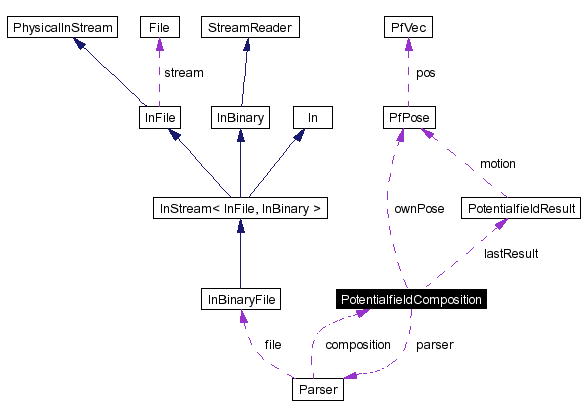
#include <PotentialfieldComposition.h>
Collaboration diagram for PotentialfieldComposition:

Public Member Functions | |
| PotentialfieldComposition () | |
| Constructor. | |
| ~PotentialfieldComposition () | |
| Destructor. | |
| void | load (const std::string &filename) |
| Loads a configuration file. | |
| void | close () |
| Destroys all objects and fields. | |
| void | execute (PotentialfieldResult &result) |
| Executes computation. | |
| void | setName (const std::string &name) |
| Sets the name. | |
| std::string | getName () const |
| Returns the name. | |
| void | setObjectState (ObjectStateDescription &desc) |
| Sets the state of one object (may set desc.objectId). | |
| void | setOwnPose (const PfPose &pose) |
| Sets the own pose. | |
| void | addField (Potentialfield *field) |
| Adds a new field to the composition. | |
| void | addObject (Object *object) |
| Adds a new object to the composition. | |
| void | addDynamicObjectState (const std::string &objectName) |
| Adds a new symbol for external object states. | |
| unsigned int | getIdFromObjectStateSymbol (const std::string &objectName) |
| Returns the internal id of a symbol. | |
| ObjectStateDescription | getDescriptionFromId (unsigned int objectId) |
| Returns a object description. | |
| std::vector< std::string > | getFieldNames () |
| Returns a list of the names of all fields. | |
| void | setFieldSelectionParameters (SelectionProcedure selectionProcedure, unsigned int n) |
| Sets parameters for field selection. | |
| void | getValueArray (const std::string &fieldname, double x1, double y1, double x2, double y2, int xSteps, int ySteps, double value[], double &max) |
| Computes an array of field values, used by visualization. | |
| void | getDirectionArray (const std::string &fieldname, double x1, double y1, double x2, double y2, int xSteps, int ySteps, PfVec directions[]) |
| Computes an array of gradient directions, used by visualization. | |
| void | setFieldActivation (const std::string &fieldname, bool activation) |
| Activates or deactivates a field. | |
Private Member Functions | |
| PotentialfieldResult | selectNextResult () |
| Selects the next result from the list. | |
| void | addResultToList (const PotentialfieldResult &result) |
| Adds a result to the list and keeps the size of the list <= n. | |
| void | updateDynamicObjects () |
| Updates the state of all dynamic objects. | |
| unsigned int | getFieldIndexFromName (const std::string &fieldname) |
| Returns the index of a field. | |
Private Attributes | |
| std::string | name |
| The name of the composition. | |
| std::vector< Object * > | objects |
| A list of all objects. | |
| NameToIndexMap | objectMap |
| Fast mapping from object names to indizes. | |
| std::vector< ObjectStateDescription > | dynamicObjectStates |
| The states of the dynamic objects. | |
| NameToIndexMap | objectStateMap |
| Fast mapping from object state names to indizes. | |
| std::vector< Potentialfield * > | fields |
| A list of all fields. | |
| NameToIndexMap | fieldMap |
| Fast mapping from field names to indizes. | |
| std::vector< PotentialfieldResult > | results |
| A list for the results of all fields. | |
| std::list< PotentialfieldResult > | resultList |
| A list for the selected results of the last runs. | |
| PotentialfieldResult | lastResult |
| The result of the last run. | |
| PfPose | ownPose |
| The own pose. | |
| Parser * | parser |
| The parser. | |
| bool | fileLoaded |
| A flag. | |
| SelectionProcedure | selectionProcedure |
| The procedure to choose the next active field. | |
| unsigned int | n |
| A parameter for the selectionProcedure. | |
Friends | |
| class | Parser |
| The PotentialfieldComposition is a friend of its parser. | |
It contains all objects and fields, executes the fields and selects the best action.
Definition at line 123 of file PotentialfieldComposition.h.
|
|
Constructor.
Definition at line 19 of file PotentialfieldComposition.cpp. References PotentialfieldResult::action, fileLoaded, and PotentialfieldResult::timeStamp. |
|
|
Destructor.
Definition at line 27 of file PotentialfieldComposition.cpp. References close(), and fileLoaded. |
Here is the call graph for this function:

|
|
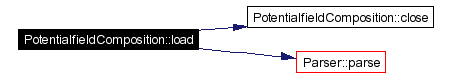
Loads a configuration file.
Definition at line 36 of file PotentialfieldComposition.cpp. References close(), fileLoaded, Parser::parse(), Parser, and parser. Referenced by GTStandardConverter::load(). |
Here is the call graph for this function:
|
|
Destroys all objects and fields.
Definition at line 49 of file PotentialfieldComposition.cpp. References dynamicObjectStates, fields, fileLoaded, and results. Referenced by load(), and ~PotentialfieldComposition(). |
|
|
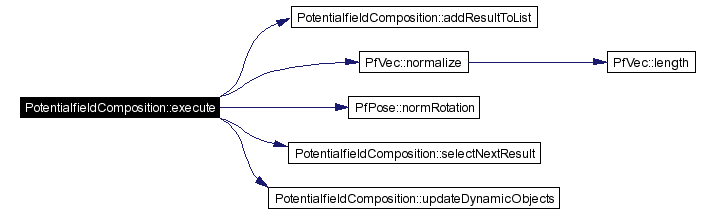
Executes computation.
Definition at line 69 of file PotentialfieldComposition.cpp. References PotentialfieldResult::action, PotentialfieldResult::actionPossible, addResultToList(), fieldMap, PotentialfieldResult::fieldNumber, fields, PotentialfieldResult::motion, PfVec::normalize(), PfPose::normRotation(), ownPose, PfPose::pos, results, PfPose::rotation, selectNextResult(), PotentialfieldResult::timeStamp, updateDynamicObjects(), and PotentialfieldResult::value. Referenced by GTStandardConverter::execute(). |
Here is the call graph for this function:
|
|
Sets the name.
Definition at line 148 of file PotentialfieldComposition.h. |
|
|
Returns the name.
Definition at line 154 of file PotentialfieldComposition.h. |
|
|
Sets the state of one object (may set desc.objectId).
Definition at line 215 of file PotentialfieldComposition.cpp. References dynamicObjectStates, getIdFromObjectStateSymbol(), ObjectStateDescription::isActive, ObjectStateDescription::objectId, ObjectStateDescription::objectName, and ObjectStateDescription::pose. Referenced by GTStandardConverter::execute(), and GTStandardConverter::setData(). |
Here is the call graph for this function:

|
|
Sets the own pose.
Definition at line 230 of file PotentialfieldComposition.cpp. References ownPose. Referenced by GTStandardConverter::setData(). |
|
|
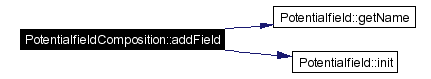
Adds a new field to the composition.
Definition at line 236 of file PotentialfieldComposition.cpp. References fieldMap, fields, Potentialfield::getName(), Potentialfield::init(), and results. Referenced by Parser::parseActionfield(), and Parser::parseMotionfield(). |
Here is the call graph for this function:
|
|
Adds a new object to the composition.
Definition at line 246 of file PotentialfieldComposition.cpp. References Object::getName(). Referenced by Parser::parseFormationObject(), and Parser::parseInstance(). |
Here is the call graph for this function:

|
|
Adds a new symbol for external object states.
Definition at line 253 of file PotentialfieldComposition.cpp. References dynamicObjectStates, ObjectStateDescription::isActive, ObjectStateDescription::objectId, ObjectStateDescription::objectName, and objectStateMap. Referenced by Parser::parseObjectStateSymbol(). |
|
|
Returns the internal id of a symbol.
Definition at line 264 of file PotentialfieldComposition.cpp. References objectStateMap. Referenced by GTStandardConverter::load(), Parser::parseInstance(), and setObjectState(). |
|
|
Returns a object description.
Definition at line 273 of file PotentialfieldComposition.cpp. References dynamicObjectStates. Referenced by Object::updateData(). |
|
|
Returns a list of the names of all fields.
Definition at line 280 of file PotentialfieldComposition.cpp. References fields. Referenced by GTStandardConverter::getFieldNames(). |
|
||||||||||||
|
Sets parameters for field selection.
Definition at line 203 of file PotentialfieldComposition.h. Referenced by Parser::parseComposition(). |
|
||||||||||||||||||||||||||||||||||||||||
|
Computes an array of field values, used by visualization.
Definition at line 309 of file PotentialfieldComposition.cpp. References fields, getFieldIndexFromName(), Potentialfield::getValueArray(), max, and updateDynamicObjects(). Referenced by GTStandardConverter::getValueArray(). |
Here is the call graph for this function:
|
||||||||||||||||||||||||||||||||||||
|
Computes an array of gradient directions, used by visualization.
Definition at line 319 of file PotentialfieldComposition.cpp. References fields, Potentialfield::getDirectionArray(), getFieldIndexFromName(), and updateDynamicObjects(). Referenced by GTStandardConverter::getDirectionArray(). |
Here is the call graph for this function:
|
||||||||||||
|
Activates or deactivates a field.
Definition at line 292 of file PotentialfieldComposition.cpp. References fields, and getFieldIndexFromName(). |
Here is the call graph for this function:
|
|
Selects the next result from the list.
Definition at line 143 of file PotentialfieldComposition.cpp. References BEST_OF_N, resultList, and selectionProcedure. Referenced by execute(). |
|
|
Adds a result to the list and keeps the size of the list <= n.
Definition at line 205 of file PotentialfieldComposition.cpp. References resultList. Referenced by execute(). |
|
|
Updates the state of all dynamic objects.
Definition at line 329 of file PotentialfieldComposition.cpp. Referenced by execute(), getDirectionArray(), and getValueArray(). |
|
|
Returns the index of a field.
Definition at line 299 of file PotentialfieldComposition.cpp. References fieldMap. Referenced by getDirectionArray(), getValueArray(), and setFieldActivation(). |
|
|
The PotentialfieldComposition is a friend of its parser.
Definition at line 241 of file PotentialfieldComposition.h. Referenced by load(). |
|
|
The name of the composition.
Definition at line 245 of file PotentialfieldComposition.h. |
|
|
A list of all objects.
Definition at line 247 of file PotentialfieldComposition.h. Referenced by Parser::getInstance(). |
|
|
Fast mapping from object names to indizes.
Definition at line 249 of file PotentialfieldComposition.h. Referenced by Parser::getInstance(). |
|
|
The states of the dynamic objects.
Definition at line 251 of file PotentialfieldComposition.h. Referenced by addDynamicObjectState(), close(), getDescriptionFromId(), and setObjectState(). |
|
|
Fast mapping from object state names to indizes.
Definition at line 253 of file PotentialfieldComposition.h. Referenced by addDynamicObjectState(), and getIdFromObjectStateSymbol(). |
|
|
A list of all fields.
Definition at line 255 of file PotentialfieldComposition.h. Referenced by addField(), close(), execute(), getDirectionArray(), getFieldNames(), getValueArray(), and setFieldActivation(). |
|
|
Fast mapping from field names to indizes.
Definition at line 257 of file PotentialfieldComposition.h. Referenced by addField(), execute(), and getFieldIndexFromName(). |
|
|
A list for the results of all fields.
Definition at line 259 of file PotentialfieldComposition.h. Referenced by addField(), close(), and execute(). |
|
|
A list for the selected results of the last runs.
Definition at line 261 of file PotentialfieldComposition.h. Referenced by addResultToList(), and selectNextResult(). |
|
|
The result of the last run.
Definition at line 263 of file PotentialfieldComposition.h. |
|
|
The own pose.
Definition at line 265 of file PotentialfieldComposition.h. Referenced by execute(), and setOwnPose(). |
|
|
The parser.
Definition at line 267 of file PotentialfieldComposition.h. Referenced by load(). |
|
|
A flag.
Definition at line 269 of file PotentialfieldComposition.h. Referenced by close(), load(), PotentialfieldComposition(), and ~PotentialfieldComposition(). |
|
|
The procedure to choose the next active field.
Definition at line 271 of file PotentialfieldComposition.h. Referenced by selectNextResult(). |
|
|
A parameter for the selectionProcedure.
Definition at line 273 of file PotentialfieldComposition.h. |
 1.3.6
1.3.6